
SCOTT Catherine
Recommendations: 0
Review: 1
Review: 1
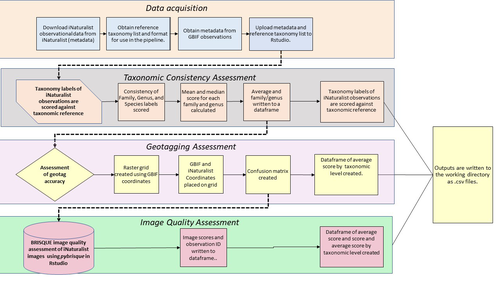
A pipeline for assessing the quality of images and metadata from crowd-sourced databases.
Harnessing the full potential of iNaturalist and other databases
Recommended by Matthias Foellmer based on reviews by Clive Hambler and Catherine ScottThe popularity of iNaturalist and other online biodiversity databases to which the general public and specialists alike contribute observations has skyrocketed in recent years (Dance 2022). The AI-based algorithms (computer vision) which provide the first identification of a given organism on an uploaded photograph have become very sophisticated, suggesting initial identifications often down to species level with a surprisingly high degree of accuracy. The initial identifications are then confirmed or improved by feedback from the community, which works particularly well for organismal groups to which many active community members contribute, such as the birds. Hence, providing initial observations and identifying observations of others, as well as browsing the recorded biodiversity for given locales or the range of occurrences of individual taxa has become a meaningful and satisfying experience for the interested naturalist. Furthermore, several research studies have now been published relying on observations uploaded to iNaturalist (Szentivanyi and Vincze 2022). However, using the enormous amount of natural history data available on iNaturalist in a systematic way has remained challenging, since this requires not only retrieving numerous observations from the database (in the hundreds or even thousands), but also some level of transparent quality control.
Billotte (2022) provides a protocol and R scripts for the quality assessment of downloaded observations from iNaturalist, allowing an efficient and reproducible stepwise approach to prepare a high-quality data set for further analysis. First, observations with their associated metadata are downloaded from iNaturalist, along with the corresponding entries from the Global Biodiversity Information Facility (GBIF). In addition, a taxonomic reference list is obtained (these are available online for many taxa), which is used to assess the taxonomic consistency in the dataset. Second, the geo-tagging is assessed by comparing the iNaturalist and GBIF metadata. Lastly, the image quality is assessed using pyBRISQUE. The approach is illustrated using spiders (Araneae) as an example. Spiders are a very diverse taxon and an excellent taxonomic reference list is available (World Spider Catalogue 2022). However, spiders are not well known to most non-specialists, and it is not easy to take good pictures of spiders without using professional equipment. Therefore, the ability of iNaturalist’s computer vision to provide identifications is limited to this date and the community of specialists active on iNaturalist is comparatively small. Hence, spiders are a good taxon to demonstrate how the pipeline results in a quality-controlled dataset based on crowed-sourced data. Importantly, the software employed is free to use, although inevitably, the initial learning curve to use R scripts can be steep, depending on prior expertise with R/RStudio. Furthermore, the approach is employable with databases other than iNaturalist.
In summary, Billotte's (2022) pipeline allows researchers to use the wealth of observations on iNaturalist and other databases to produce large metadata and image datasets of high-quality in a reproducible way. This should pave the way for more studies, which could include, for example, the assessment of range expansions of invasive species or the evaluation of the presence of endangered species, potentially supporting conservation efforts.
References
Billotte J (2022) A pipeline for assessing the quality of images and metadata from crowd-sourced databases. BiorXiv, 2022.04.29.490112, ver 5 peer reviewed and recommended by Peer Community In Zoology. https://doi.org/10.1101/2022.04.29.490112
Dance A (2022) Community science draws on the power of the crowd. Nature, 609, 641–643. https://doi.org/10.1038/d41586-022-02921-3
Szentivanyi T, Vincze O (2022) Tracking wildlife diseases using community science: an example through toad myiasis. European Journal of Wildlife Research, 68, 74. https://doi.org/10.1007/s10344-022-01623-5
World Spider Catalog (2022). World Spider Catalog. Version 23.5. Natural History Museum Bern, online at http://wsc.nmbe.ch. https://doi.org/10.24436/2