
Harnessing the full potential of iNaturalist and other databases
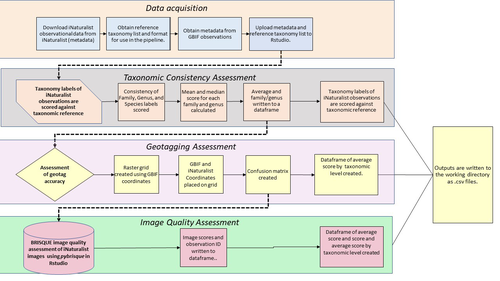
A pipeline for assessing the quality of images and metadata from crowd-sourced databases.
Abstract
Recommendation: posted 11 November 2022, validated 30 November 2022
Foellmer, M. (2022) Harnessing the full potential of iNaturalist and other databases. Peer Community in Zoology, 100017. https://doi.org/10.24072/pci.zool.100017
Recommendation
The popularity of iNaturalist and other online biodiversity databases to which the general public and specialists alike contribute observations has skyrocketed in recent years (Dance 2022). The AI-based algorithms (computer vision) which provide the first identification of a given organism on an uploaded photograph have become very sophisticated, suggesting initial identifications often down to species level with a surprisingly high degree of accuracy. The initial identifications are then confirmed or improved by feedback from the community, which works particularly well for organismal groups to which many active community members contribute, such as the birds. Hence, providing initial observations and identifying observations of others, as well as browsing the recorded biodiversity for given locales or the range of occurrences of individual taxa has become a meaningful and satisfying experience for the interested naturalist. Furthermore, several research studies have now been published relying on observations uploaded to iNaturalist (Szentivanyi and Vincze 2022). However, using the enormous amount of natural history data available on iNaturalist in a systematic way has remained challenging, since this requires not only retrieving numerous observations from the database (in the hundreds or even thousands), but also some level of transparent quality control.
Billotte (2022) provides a protocol and R scripts for the quality assessment of downloaded observations from iNaturalist, allowing an efficient and reproducible stepwise approach to prepare a high-quality data set for further analysis. First, observations with their associated metadata are downloaded from iNaturalist, along with the corresponding entries from the Global Biodiversity Information Facility (GBIF). In addition, a taxonomic reference list is obtained (these are available online for many taxa), which is used to assess the taxonomic consistency in the dataset. Second, the geo-tagging is assessed by comparing the iNaturalist and GBIF metadata. Lastly, the image quality is assessed using pyBRISQUE. The approach is illustrated using spiders (Araneae) as an example. Spiders are a very diverse taxon and an excellent taxonomic reference list is available (World Spider Catalogue 2022). However, spiders are not well known to most non-specialists, and it is not easy to take good pictures of spiders without using professional equipment. Therefore, the ability of iNaturalist’s computer vision to provide identifications is limited to this date and the community of specialists active on iNaturalist is comparatively small. Hence, spiders are a good taxon to demonstrate how the pipeline results in a quality-controlled dataset based on crowed-sourced data. Importantly, the software employed is free to use, although inevitably, the initial learning curve to use R scripts can be steep, depending on prior expertise with R/RStudio. Furthermore, the approach is employable with databases other than iNaturalist.
In summary, Billotte's (2022) pipeline allows researchers to use the wealth of observations on iNaturalist and other databases to produce large metadata and image datasets of high-quality in a reproducible way. This should pave the way for more studies, which could include, for example, the assessment of range expansions of invasive species or the evaluation of the presence of endangered species, potentially supporting conservation efforts.
References
Billotte J (2022) A pipeline for assessing the quality of images and metadata from crowd-sourced databases. BiorXiv, 2022.04.29.490112, ver 5 peer reviewed and recommended by Peer Community In Zoology. https://doi.org/10.1101/2022.04.29.490112
Dance A (2022) Community science draws on the power of the crowd. Nature, 609, 641–643. https://doi.org/10.1038/d41586-022-02921-3
Szentivanyi T, Vincze O (2022) Tracking wildlife diseases using community science: an example through toad myiasis. European Journal of Wildlife Research, 68, 74. https://doi.org/10.1007/s10344-022-01623-5
World Spider Catalog (2022). World Spider Catalog. Version 23.5. Natural History Museum Bern, online at http://wsc.nmbe.ch. https://doi.org/10.24436/2
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
No indication
Evaluation round #2
DOI or URL of the preprint: https://doi.org/10.1101/2022.04.29.490112
Version of the preprint: 2
Author's Reply, 18 Oct 2022
Decision by Matthias Foellmer, posted 05 Sep 2022
Dear Ms. Billotte,
Thank you for addressing the reviewers’ comments in such a thorough manner. I have only a few comments left regarding the writing/presentation:
L 9/10: avoid repetitions
L42: delete comma after “image-based”
L63/64: switch “the of” to “of the”
Figure 2: The labeling seems incomplete. In the legend, you refer to Section 1, 2, and 3. Which sections are these?
Figure 3: In the legend, last sentence, it should be “observations” (plural).
Results: please check the numbers one more time, or at least clarify. On L171, you state that you found 156,842 downloadable observations and on L176, you say that 49.91% were identified to at least family level. But on L181, you state that 156,842 out of 158,129 downloadable observations had a family-level identification. On L185, you refer to 425,950 “records”. What is a record in this context, i.e. how does it differ from an observation?
L223-225: use “research grade” throughout.
L234: delete comma after “quantifiable”
Zizka et al. 2019 is not in the reference list.
Kind regards,
Matthias Foellmer, NYC, 5 September 2022
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2022.04.29.490112
Version of the preprint: 1
Author's Reply, 18 Aug 2022
Decision by Matthias Foellmer, posted 22 Jun 2022
Dear Jackie Bilotte,
In this manuscript you present a protocol to efficiently evaluate the quality of images and associated metadata of any pre-specified taxon using self-written and co-opted R and MATLAB scripts. Importantly, the usefulness of the approach extends to e.g. assessing the range expansions of invasive species or evaluating the presence of endangered species, potentially supporting conservation efforts. Given the rapidly increasing popularity of iNaturalist and similar databases to which the general public contributes, this paper can make a very valuable and timely contribution. The reviewers agree with this view and provide thoughtful suggestions for improving the clarity and facilitating the use of the various workflow steps. It would be fantastic if you could make BRISQUE work in Python, since, as one of the reviewers points out, few readers will probably have access to MATLAB, which is expensive.
In additions to the reviewers’ comments, I have the following suggestions:
L20: “… but lower in observations…” – wouldn’t “for” be more appropriate than “in”?
Data acquisition:
- You state that “For the Araneae case study, I searched for and downloaded observations for each family under the order Araneae on iNaturalist on July 21, 2021 (‘iNaturalist’, 2014). I then searched for and downloaded observations classified only to the order level.” Please explain why you employed this strategy. One would think that a single query for Araneae should give all relevant results, with all observations determined to the various taxonomic levels. After all, downloading data for 100+ families one-by-one seems an arduous endeavor I would want to avoid.
- Datasets to be downloaded from iNat can easily be very large without narrowing down the search criteria. Please detail your search strategy for at least one example. Specify the settings in the filter and the Export Observations page, so that the reader can reproduce your search results (see also the reviewers’ comments). On lines 87ff you only state the minimum requirements with respect to the fields to be included.
- When I tried to run your R code for obtaining the data directly from iNat for taxon_name = "Araneidae", I got the error message “Error in get_inat_obs(query = NULL, taxon_name = "Araneidae", taxon_id = NULL, : Your search returned too many results, please consider breaking it up into smaller chunks by year or month.” So simply searching for a given family doesn’t work, highlighting the need for a more detailed description of your search strategy.
- Your site Observation_Database_Assesment on GitHub.com currently (when I checked) only has the basic R and MATLAB code posted, but no other files. Please add example searches and data sets.
R and MATLAB code: please make sure you provide sufficient annotation so that all steps and their implementation can easily be understood even by the not-so-proficient coder.
I hope you find the reviewers’ and my comments helpful. I’m looking forward to reading your revision.
Matthias Foellmer, NYC, 22-Jun-2022
Reviewed by Catherine Scott, 06 Jun 2022
This is a valuable and timely contribution. As an arachnologist interested in using iNaturalist data, I think it provides some very useful tools and guidelines for processing and using these data.
Major comments:
Unlike R, which is freely available, MATLAB is a paid software so it is not accessible to all. If possible, it would be preferable to have a script for the image analysis in a freely available software. It seems that BRISQUE can be implemented in Python: https://github.com/bukalapak/pybrisque. I do really like the suggestion that iNaturalist automatically score image quality--this would make filtering out useful observations much easier!
While not critical for this paper, which is meant to describe methods, it would be really nice to have an example of the utility of some of the methods, perhaps for a particular family (one of the smaller ones). Going through each of the steps and making the specific small dataset and code available for readers to reproduce the analyses to familiarize themselves with the pipeline, knowing the expected results, would be very valuable. It would also be good to show an example where the GBIF and iNAT ranges do not match, and a look at whether it's because of a mislabeled observation or a true range expansion.
Minor comments:
since data were non-Normal, it might be more appropriate and informative to report medians and ranges rather than means and SE
Figure 3 caption is cut off
line 162: Araneidae must be a typo--presumably this should be one of the smaller families that start with A
line 168: "I found 158,129 of the 156,842 downloadable observations" check numbers, have they been reversed?
line 173: it would be helpful to have some explanation of what it means for an observation to be accurate or precise.
line 206: it is not quite correct that "requires that an observation reach a threshold of three votes from users to confirm an identification" as research-grade. Instead, iNaturalist states that "Observations become "Research Grade" when the community agrees on species-level ID or lower, i.e. when more than 2/3 of identifiers agree on a taxon." In practice this means that an observation can become research grade after exactly two identifiers agree on an ID.
Note: I did not have a chance to try to run any of the code myself.
https://doi.org/10.24072/pci.zool.100128.rev11









