
Multi-stress responses depend on the microbiome in the planktonic crustacean Daphnia
Microbiome mediated tolerance to biotic stressors: a case study of the interaction between a toxic cyanobacterium and an oomycete-like infection in Daphnia magna
Abstract
Recommendation: posted 21 April 2023, validated 26 April 2023
Visser, B. and Scheifler, M. (2023) Multi-stress responses depend on the microbiome in the planktonic crustacean Daphnia. Peer Community in Zoology, 100110. https://doi.org/10.24072/pci.zool.100110
Recommendation
The critical role that gut microbiota play in many aspects of an animal’s life, including pathogen resistance, detoxification, digestion, and nutritional physiology, is becoming more and more apparent (Engel and Moran 2013; Lindsay et al., 2020). Gut microbiota recruitment and maintenance can be largely affected by the surrounding environment (Chandler et al., 2011; Callens et al., 2020). The environment may thus dictate gut microbiota composition and diversity, which in turn can affect organismal responses to stress. Only few studies have, however, taken the gut microbiota into account to estimate life histories in response to multiple stressors in aquatic systems (Macke et al., 2016).
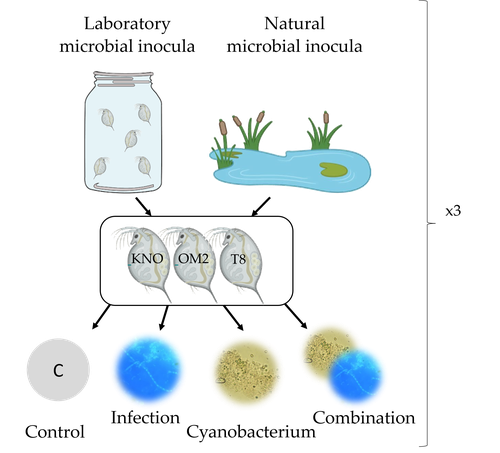
Houwenhuyse et al., investigate how the microbiome affects life histories in response to ecologically relevant single and multiple biotic stressors (an oomycete-like parasite, and a toxic cyanobacterium) in Daphnia magna (Houwenhuyse et al., 2023). Daphnia is an excellent model, because this aquatic system lends itself extremely well for gut microbiota transplantation and manipulation. This is due to the possibility to sterilize eggs (making them free of bacteria), horizontal transmission of bacteria from the environment, and the relative ease of culturing genetically similar Daphnia clones in large numbers.
The authors use an elegant experimental design to show that the Daphnia gut microbial community differs when derived from a laboratory versus natural inoculum, the latter being more diverse. The authors subsequently show that key life history traits (survival, fecundity, and body size) depend on the stressors (and combination thereof), the microbiota (structure and diversity), and Daphnia genotype. A key finding is that Daphnia exposed to both biotic stressors show an antagonistic interaction effect on survival (being higher), but only in individuals containing laboratory gut microbiota. The exact mechanism remains to be determined, but the authors propose several interesting hypotheses as to why Daphnia with more diverse gut microbiota do less well. This could be due, for example, to increased inter-microbe competition or an increased chance of contracting opportunistic, parasitic bacteria. For Daphnia with less diverse laboratory gut microbiota, a monopolizing species may be particularly beneficial for stress tolerance. Alongside these interesting findings, the paper also provides extensive information about the gut microbiota composition (available in the supplementary files), which is a very useful resource for other researchers.
Overall, this study reveals that multiple, interacting factors affect the performance of Daphnia under stressful conditions. Of importance is that laboratory studies may be based on simpler microbiota systems, meaning that stress responses measured in the laboratory may not accurately reflect what is happening in nature.
REFERENCES
Callens M, De Meester L, Muylaert K, Mukherjee S, Decaestecker E. The bacterioplankton community composition and a host genotype dependent occurrence of taxa shape the Daphnia magna gut bacterial community. FEMS Microbiology Ecology. 2020;96(8):fiaa128. https://doi.org/10.1093/femsec/fiaa128
Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLOS Genetics. 2011;7(9):e1002272. https://doi.org/10.1371/journal.pgen.1002272
Engel P, Moran NA. The gut microbiota of insects - diversity in structure and function. FEMS Microbiology Reviews. 2013;37(5):699-735. https://doi.org/10.1111/1574-6976.12025
Houwenhuyse S, Bulteel L, Vanoverberghe I, Krzynowek A, Goel N et al. Microbiome mediated tolerance to biotic stressors: a case study of the interaction between a toxic cyanobacterium and an oomycete-like infection in Daphnia magna. 2023. OSF, ver. 2 peer-reviewed and recommended by Peer Community in Zoology. https://doi.org/10.31219/osf.io/9n4mg
Lindsay EC, Metcalfe NB, Llewellyn MS. The potential role of the gut microbiota in shaping host energetics and metabolic rate. Journal of Animal Ecology. 2020;89(11):2415-2426. https://doi.org/10.1111/1365-2656.13327
Macke E, Tasiemski A, Massol F, Callens M, Decaestecker E. Life history and eco-evolutionary dynamics in light of the gut microbiota. Oikos. 2017;126(4):508-531. https://doi.org/10.1111/oik.03900
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
Funding was provided by the KU Leuven research project C16/17/002; ID-N PlasticDaphnia and the FWO projects G092619N and G06216N.
Evaluation round #3
DOI or URL of the preprint: https://doi.org/10.31219/osf.io/3vgdh
Version of the preprint: 2
Author's Reply, 04 Apr 2023
Decision by Bertanne Visser and Mathilde Scheifler , posted 30 Jan 2023, validated 30 Jan 2023
, posted 30 Jan 2023, validated 30 Jan 2023
Dear authors,
Thanks for submitting the revision of your paper. Together, we have reviewed your paper and have made several minor suggestions that we think will improve the paper (included as track changes and comments in your word file). We would be happy to recommend your paper following these minor revisions.
Thanks in advance. Yours sincerely,
Bertanne Visser and Mathilde Scheifler
Download recommender's annotationsEvaluation round #2
DOI or URL of the preprint: https://doi.org/10.31219/osf.io/ws7m6
Version of the preprint: 1
Author's Reply, 15 Dec 2022
Decision by Bertanne Visser, posted 16 Nov 2021
Minor revisions before recommendation
Dear authors,
Thanks for your detailed response to the reviewers and your revision. Please find attached your revision with several minor comments. Please address these minor comments for a final revision. I will then be happy to recommend your paper for PCI Zoology.
Yours sincerely,
Bertanne Visser
Download recommender's annotationsEvaluation round #1
DOI or URL of the preprint: https://doi.org/10.31219/osf.io/3vgdh
Version of the preprint: 1
Author's Reply, 09 Oct 2021
Decision by Bertanne Visser, posted 19 Jul 2021
Dear authors,
I have now received the reports of three reviewers. Overall, all reviewers were positive about your pre-print and I agree with their assessments. The reviewers made several suggestions that I think would improve your manuscript before recommendation. Please revise your manuscript accordingly and address each of the reviewer suggestions in your rebuttal.
I look forward to receiving your revision.
Yours sincerely,
Bertanne Visser
Reviewed by Natacha Kremer, 08 Jul 2021
In this paper, the authors test the influence of water (laboratory vs. natural) used for the colonization of axenic Daphnia magna (3 different genotypes) on their response to two biotic stressors: a toxic cyanobacterium (Microcystis aeruginosa) and a pathogenic fungus (Aspergillus aculeatus like). They test how these biotic stressors, alone or in combination, impact host life-history traits and modify the gut microbiota composition. They show that gut microbiota diversity somehow mirrors the diversity of the bacterial composition of the ‘inoculation’ water (i.e., more diverse community in Daphnia inoculated with water from natural pounds that also have a higher diversity). An interaction between the ‘inoculation’ water and the response against stressors occurs for survival, but not for the other tested life history traits (fecundity and body size). Bacterial composition was not strongly impacted by stressors.
This study is very interesting and points out how laboratory conditions can potentially bias the response to stressors. In addition, it considers the combination of stressors, which is generally lacking in this field. However, a few points need to be considered:
- The study is nicely designed but 3-factors analyses are complex. The presentation of results is complicated to follow and would benefit being clarified.
- For microbial analyses, various sampling efforts may bias the analyses and interpretation.
- Results should be interpreted and discussed more in light of the ‘colonization then stress’ experimental sequence and of the dynamics of colonization rather than a concomitant selective process.
- In addition, it would be interesting to address the question of bacterial density (and noy only diversity) in this system (and its potential cost).
All these points are detailed bellow.
General suggestions for reorganization / clarification:
- Move the M&M before the results, otherwise, it is not possible to understand the experiments
- Present Aspergillus (and potentially the last paragraph of the results) in the M&M (model system)
- Restructure the part linked to the model system: 1) to combine information on feeding in the same paragraph, 2) present all stressor agents and their phenotypic effect together
- Add an extra scheme in the M&M indicating the sequence of the experiment (germ-free -> colonization -> stress)
- Always use the same nomenclature for the experimental conditions (they are different between analyses of life-history traits and microbiota). In addition, ‘infection’ is misleading because both the treatments (cyanobacterium and fungi) are biotic stressors. I would rather use ‘control’ (= Ctl), ‘cyanobacterium’ (= C)(or ‘Microcystis’), ‘fungus’ (= F) (or ‘Aspergillus’), ‘combination’ (=C+F)
- Maybe you could structure the result section with the scientific questions you are asking and not the measured traits.
- Do not start the result section with the table of result
- Union plots are difficult to read (for me who have never seen this type of graphs before). At minimum, it would be nice to see the categories better (with a cartoon for each stress?)
- I would start the description of bacterial diversity with OTU richness and beta-diversity (and not necessarily present the union plots)
- L517-529: this part seems to be a ‘result’ part.
- I did not have access to the supplementary tables
Introduction
-l22-24: This sentence is not very clear and does not seem to be supported by the results (no difference observed in the microbiome whatever the stressor condition)
-l55: the production of proteases or genes involved in secondary metabolic transport and catabolism do not seem to be ‘defence’ mechanisms against Microcystis (as they do not participate to its elimination), but rather ‘tolerance’ mechanisms (as they limit the cost of their presence)
- l88-90: Are the variation in gut communities reflecting a selective filtration process or just a random uptake of bacteria from the environment? Is there any order of colonization (with pioneer species)? Is the dynamic of colonization within Daphnia gut already known?
-l110: please define tolerance in this context
-l123-125: Is the overall/specific bacterial load higher in natural pounds than in lab water? In colonized Daphnia, do we observe variation in bacterial load in the gut microbiota (and a cost associated with higher loads)?
Results:
- Table 1: I don’t understand to what refers the ‘sample type’ in the table
- Table 1 and all over the manuscript: ‘Donor’ and ‘Recipient’ are used in different ways. Sometimes it seems to be the water that has been used for inoculation (from ponds or water tanks where Daphnia were reared) but sometimes, is seems to be the Daphnia from the tanks (and not the water). In the table, I would be more explicit using terms such as ‘water used for inoculation’, ‘Daphnia + Microbiota’, and throughout the text ‘ex: l204, l261, l274, l281, l537), I would indicate that you refer to the water and not to donor Daphnia. By the way, is the community from donor Daphnia very different from the one in the water in which they are reared?
- Fig 1: As 3 replicates are used, could you indicate the variance associated with these curves?
- Fig 1: Condition microbiome control / genotypes KNO and T8: the control treatment exhibits a lower survival than the stressors treatments. Can we thus talk about ‘stressors’? Is it a cost of harboring gut symbionts (at a higher density?)?
- Fig 2: how do you explain such variance in control treatments?
- L216: Union plots do not indicate the link between gut microbial communities and life-history traits but between gut microbial communities and stressors. In addition, why to go into detail since the stressors did not impact the bacterial community in daphnia (table 1: OTU richness and Beta diversity)? Finally, what is the significance between the numbers in each class?
- Fig 4: please indicate the meaning of the letters (and homogenize the nomenclature). Why is the number of replicates different in the different combination genotype x stressor * environment? How do you explain the high variability in some samples (KNO/Lab/M2 or OM2/lab/F2)? Please also indicate the composition of the water used for inoculation.
- Fig5: please make these graphs more easily readable. Do they represent the combination of the 3 replicates?
- Fig S5: The number or samples is very different between donor and recipient communities. Could the sampling effort bias the analysis? (e.g., if we pick randomly 3 green dots and 3 blue dots from the PCA from recipient communities, there is a high chance that they structure separately as they do for the donor community).
- Fig 11: the number of replicates seems different between conditions (as in fig4). Could this impact the interpretation of the structuration of bacterial communities?
- The characterization of the infection by Aspergillus does not really fit to the general question, but more to the characterization of the stressor (method)
Discussion:
- L362: Is the difference in one OTU when all replicate samples are merged per condition?
- L366: ‘selection of bacterial groups was stressor-dependent’ -> If I well understood, colonization occurs during the first 2 days and only then the stressor is added. It would be interesting to modulate this sentence with the way the establishment of the colonization occurs (selection by the host might be done at 2 days, but resistance of bacteria could be different depending on the presence/absence of stressors)
- L367-368: suggest an explanation?
- It would be interesting to discuss more why survival (and indirectly fecundity) are impacted by the microbiome, while bacterial diversity and richness did not change much in response to stressors.
Methods:
- Fig12: please add 10µM before ‘filtered’, otherwise we think that the water is sterile
- Fig13: please indicate the scale bar on the image (not only the magnification)
- L630: after a first event of colonization of the gut, is a colonization by other bacteria still possible?
- Is the same ‘inoculation water’ used for the 3 replicates?
- L652: please indicate on fig4 how many guts were dissected per samples
- L655: How were designed the nested primers? Could they bias the amplification and/or limit the number of orders amplified?
- L660: why did you choose the V4 region?
- L671: why n=6, n=2 and n=1?
- L699: which samples have less than 1000 reads?
- L703: why to choose 1000 reads? Please show the rarefaction curves.
Typos / grammar:
-l22: fecundity
-l120-125: please cut your sentence in two
-l128: please remove the comma after individuals
-l240: Fig 6B
-l241: fig 6A
-l529: fig please specify to what refer ‘this’
-l733: characterization of the infection
-l741: please specify what are the LSU (=large subunit) and SSU (=small subunit) regions
https://doi.org/10.24072/pci.zool.100110.rev11
Reviewed by anonymous reviewer 2, 07 Jul 2021
Reviewed by anonymous reviewer 1, 08 Jul 2021
This paper aims to compare the effect of multiple versus single stressors on aquatic invertebrates (Daphnia magma). Emphasizing that studies on stressors are often performed under laboratory conditions, the authors contrasted Daphnia with natural and laboratory microbiota to examine their response to the stressors. They also investigated the effect of genotype. The two stressors used, in combination and individually, are a toxic cyanobacterium (Microcystis aeruginosa) and a fungal infection (Aspergillus aculeatus-like type). The effects of those stressors were determined by measuring survival, reproduction, body size, as well as the correlation with microbial diversity. This paper includes a lot of measurements and the results are thoroughly described.
Major comments:
The paper would become more interesting for a more general audience if the authors would put their work in a broader framework, considering also the work of others and the work on different systems.
The main hypotheses and the goal of the study do not become very clear from the introduction (see minor comments). Was the initial goal of the study to investigate stress responses or to know the differences between lab and natural settings or both? And why is this relevant for your system or of interest in general?
Your explicit expectations do not become very clear for the stress responses. In what direction do you expect to see differences in life history traits or microbiome diversity?
When introducing stress responses, it would be useful if the authors could add a paragraph with more concrete examples and findings in Daphnia (also to explain why this model was chosen compared to other model systems in this context). While a lot of papers on Daphnia are cited, how these works differ from the current work does not become very clear. It might be worth to mention explicitly what the novelty of this paper is compared to the broader literature (and perhaps include a full paragraph on stress responses in other organisms and why it is important).
In the introduction, you emphasize that the effect of stressors can be synergetic, additive or antagonist and that it varies according to the ecosystem (freshwater or marine), yet you never mention that Daphnia is a freshwater organism.
The results section is currently placed before the materials and methods, but as it stands, the materials and methods must be read before the results can be understood and interpreted correctly. I suggest that the authors either rewrite the results and state more clearly what was done and why for each section or that the materials and methods are placed before the results section.
Throughout the paper, there is some redundancy and repetitiveness. I have highlighted several places in the minor comments. The discussion is also largely a repetition of the main results. As mentioned above for the introduction, I think that this requires some rewriting to put the work into a broader context.
There are many figures, which reduces the coherence and readability of the paper. It would be a good idea to provide some basic stats (means, standard deviations) for the different traits measured in a table (this would already reduce the number of figures). For the remaining figures, please also add the relevant statistics and sample sizes.
Please find some minor comments in the provided PDF.
Download the review https://doi.org/10.24072/pci.zool.100110.rev13









